viral genatics
Modifications (genetics)

Phenotype mixing
Phenotype mixing is a form of interaction between two virus particles, each of which holds its own unique genetic material. The two particles "share" coat proteins, therefore each has a similar assortment of identifying surface proteins, while having different genetic material. In other words; nongenetic interaction in which virus particles released from a cell that is infected with two different viruses have components from both the infecting agents, but with a genome from one of them.[1]
MUTANTS
Origin
Spontaneous mutations
These arise naturally during viral replication: e.g. due to errors by the genome-replicating polymerase or a a result of the incorporation of tautomeric forms of the bases
Tautomers are constitutional isomers of organic compounds that readily interconvert. This reaction commonly results in the relocation of a proton. ... The chemical reaction interconverting the two is called tautomerization.
DNA viruses tend to more genetically stable than RNA viruses. There are error correction mechanisms in the host cell for DNA repair, but probably not for RNA.
Some RNA viruses are remarkably invariant in nature. Probably these viruses have the same high mutation rate as other RNA viruses, but are so precisely adapted for transmission and replication that fairly minor changes result in failure to compete successfully with parental (wild-type, wt) virus.
Mutations that are induced by physical or chemical means
Chemical:
Agents acting directly on bases, e.g. nitrous acid
Agents acting indirectly, e.g. base analogs which mispair more frequently than normal bases thus generating mutationsPhysical:
Agents such as UV light or X-rays
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is lost during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome.[1]

insertion mutation) is the addition of one or more nucleotide base pairs into a DNA sequence. This can often happen in Microsatellite regions due to the DNA polymerase slippage. Insertions can be anywhere in size from one base pair incorrectly inserted into a DNA sequence to a section of one chromosome inserted into another.
EXCHANGE OF GENETIC MATERIAL
Recombination
Exchange of genetic information between two genomes.
"Classic" recombination
This involves breaking of covalent bonds within the nucleic acid, exchange of genetic information, and reforming of covalent bonds.
This kind of break/join recombination is common in DNA viruses or those RNA viruses which have a DNA phase (retroviruses). The host cell has recombination systems for DNA.
Recombination of this type is very rare in RNA viruses (there are probably no host enzymes for RNA recombination). Picornaviruses show a form of very low efficiency recombination. The mechanism is not identical to the standard DNA mechanism, and is probably a "copy choice" kind of mechanism (figure 1) in which the polymerase switches templates while copying the RNA.
ret·ro·vi·rusˈretrōˌvīrəs/nounBIOLOGY
A retrovirus is a single-stranded positive-sense RNA virus with a DNA intermediate and, as an obligate parasite, targets a host cell. Once inside the host cell cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro(backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, translating and transcribing the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus. It is difficult to detect the virus until it has infected the host. At that point, the infection will persist indefinitely.
Reassortment
If a virus has a segmented genome and if two variants of that virus infect a single cell, progeny virions can result with some segments from one parent, some from the other.
This is an efficient process - but is limited to viruses with segmented genomes - so far the only human viruses characterized with segmented genomes are RNA viruses e.g. orthomyxoviruses, reoviruses, arenaviruses, bunya viruses.
Reassortment may play an important role in nature in generating novel reassortants and has also been useful in laboratory experiments (figure 3). It has also been exploited in assigning functions to different segments of the genome. For example, in a reassorted virus if one segment comes from virus A and the rest from virus B, we can see which properties resemble virus A and which virus B.
Reassortment is a non-classical kind of recombination
Reassortment is the mixing of the genetic material of a species into new combinations in different individuals. Several different processes contribute to reassortment, including assortment of chromosomes, and chromosomal crossover.[1] It is particularly used when two similar viruses that are infecting the same cell exchange genetic material. In particular, reassortment occurs among influenza viruses, whose genomes consist of eight distinct segments of RNA. These segments act like mini-chromosomes, and each time a flu virus is assembled, it requires one copy of each segment.
If a single host (a human, a chicken, or other animal) is infected by two different strains of the influenza virus, then it is possible that new assembled viral particles will be created from segments whose origin is mixed, some coming from one strain and some coming from another. The new reassortant strain will share properties of both of its parental lineages.
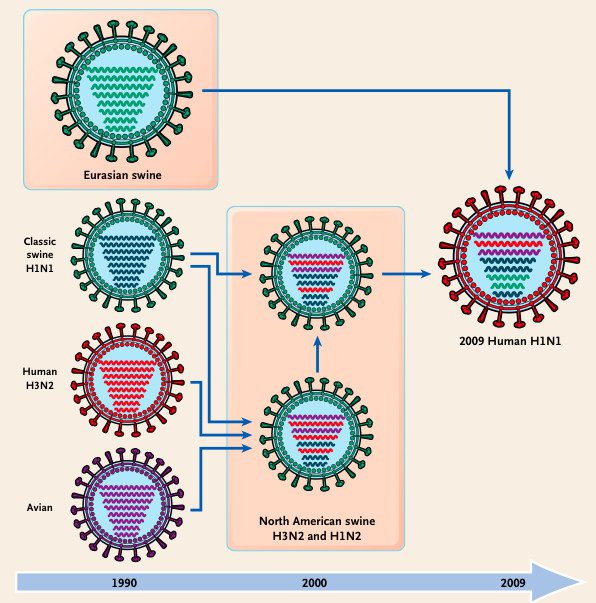
Reassortment is responsible for some of the major genetic shifts in the history of the influenza virus. The 1957 and 1968 pandemic flu strains were caused by reassortment between an avian virus and a human virus, whereas the H1N1virus responsible for the 2009 swine flu outbreak has an unusual mix of swine, avian and human influenza genetic sequences
Reassortment of the influenza virus genome
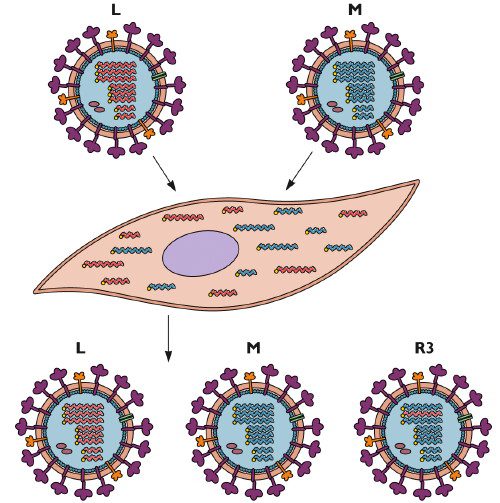
Mutation is an important source of RNA virus diversity that is made possible by the error-prone nature of RNA synthesis. Viruses with segmented genomes, such as influenza virus, have another mechanism for generating diversity: reassortment.
When an influenza virus infects a cell, the individual RNA segments enter the nucleus. There they are copied many times to form RNA genomes for new infectious virions. The new RNA segments are exported to the cytoplasm, and then are incorporated into new virus particles which bud from the cell.
If a cell is infected with two different influenza viruses, the RNAs of both viruses are copied in the nucleus. When new virus particles are assembled at the plasma membrane, each of the 8 RNA segments may originate from either infecting virus. The progeny that inherit RNAs from both parents are called reassortants. This process is illustrated in the diagram below, which shows a cell that is co-infected with two influenza viruses L and M. The infected cell produces both parental viruses as well as a reassortant R3 which inherits one RNA segment from strain L and the remainder from strain M.
One example of the evolutionary importance of reassortment is the exchange of RNA segments between mammalian and avian influenza viruses that give rise to pandemic influenza. For example, the 2009 H1N1 pandemic strain is a reassortant of avian, human, and swine influenza viruses, as illustrated.
Reassortment can only occur between influenza viruses of the same type. Why influenza A viruses never exchange RNA segments with influenza B or C viruses is not understood. However, the reason is probably linked to the packaging mechanism that ensures that each influenza virion contains at least one copy of each RNA segment.
Trifonov, V., Khiabanian, H., & Rabadan, R. (2009). Geographic Dependence, Surveillance, and Origins of the 2009 Influenza A (H1N1) Virus New England Journal of Medicine DOI: 10.1056/NEJMp0904572
- Recombination
- exchange of genes between 2 chromosomes
- crossing over occurs in homologous regions
- viruses with segmented genomes exchange segments
- analagous to high-frequency recombination
- results in influenza pandemics
- 2 viruses infect a cell, but one is mutated and has a non-functional protein
- the nonmutated virus helps the mutant by making protein for both viruses
- 2 viruses infect a cell
- virus A has its own genetic material, but the surface proteins of virus B
- progeny viruses contain coat components and genetic material from virus A
- this is because they only had the genetic material from virus A
- genetic material is not altered
- pseudovirion formation
- when a virus's coat is entirely from another virus
- nucleic acid and coat are completely mismatched




0 تعليقات